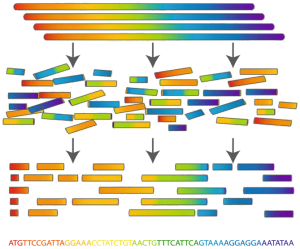
Genome assembly
In bioinformatics, genome assembly represents the process of combining many short DNA sequences to recreate the original chromosomes from which the DNA originated. Sequence assembly is one of the basic steps after performing DNA sequencing. The established genome assembly can be submitted to databases such as the European Nucleotide Archive, NCBI Genome, and Ensembl Genomes. You can also browse these databases for genomic sequences done by other researchers.
Credit: Dodona
There are two different types of genome assembly: de novo assembly and mapping to a reference genome. De novo assembly refers to the genome assembly of a novel genome from scratch without reference to genomic data. A reference genome or a reference assembly is a digital nucleic acid sequence database that represents a species’ set of genes. Once the reference genome is available, with its aid, the genome assembly becomes much easier, quicker, and even more accurate. Therefore, unless necessary, researchers choose the method of reference-based alignment. Reference-based alignment has become the current standard in diagnostics.
A high-quality and well-annotated genome assembly is increasingly becoming an essential tool for applied and basic research across many biological disciplines in the 21st century that can turn any organism into a model organism. Thus, securing complete and accurate reference genomes and annotations before analyzing post-genome studies such as genome-wide association studies, structural variations, and posttranslational studies (methylation or histone modification) has become a cornerstone of modern genomics. However, early works have warned against its applications in genome assembly because the resultant assemblies may contain biases toward errors and chromosomal rearrangements in the existing reference genome.
Additional resources
- Jung, H., Ventura, T., Chung, J. S., Kim, W. J., Nam, B. H., Kong, H. J., … & Eyun, S. I. (2020). Twelve quick steps for genome assembly and annotation in the classroom. PLoS computational biology, 16(11), e1008325. doi: 10.1371/journal.pcbi.1008325
- EE 372: Data Science for High-Throughput Sequencing